//출처 : https://en.wikipedia.org/wiki/Sequence_alignment
Sequence alignment - Wikipedia
From Wikipedia, the free encyclopedia Jump to navigation Jump to search Process in bioinformatics that identifies equivalent sites within molecular sequences In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protei
en.wikipedia.org
*Global Alignment <-> Local Alignment *
Differences
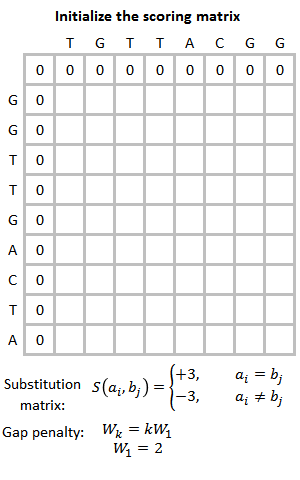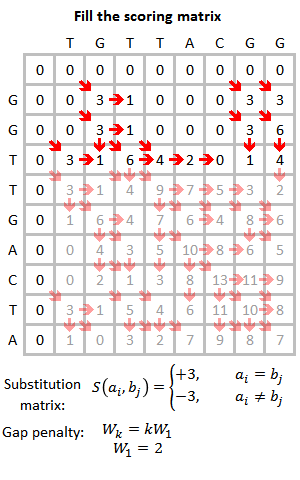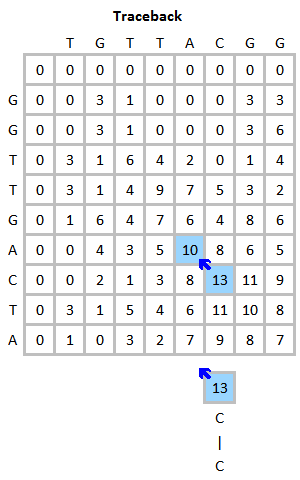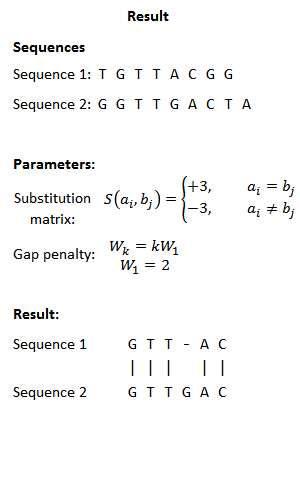
1. Build Scoring Matrix Step => if m[i][j]<0 then m[i][j] = 0
2. Back tracking Step => From Max to Zero
Global Alignment 와의 차이는 Scoring Matrix 에서 m[i][j]가 0보다 작은 값이면
0으로 채운다. 또한, Back Tracking 과정에서 Global Alignment 는 문자열의 길이만큼이 Alignment 가 되기 때문에
m[seq1.length][seq2.length] 부터 i,j가 0일때까지 back track하지만
Local Alignment 는 일치하는 값만 출력시키기 때문에 가장 큰 값에서부터, 0까지 출력시킨다.
파일 입출력을 포함해 Alignment 를 txt파일 형식으로 출력시키는 코드를 짜보았다.
#Java Source Code
import java.io.BufferedReader;
import java.io.BufferedWriter;
import java.io.File;
import java.io.FileReader;
import java.io.FileWriter;
import java.io.IOException;
public class LocalAlignment {
static String seq1="";
static String seq2="";
static String seq1align = "";
static String seq2align = "";
static int m[][];
static int trace[][];
static int digonal;
static int up;
static int left;
static int max = 0;
public static void main(String args[]) throws IOException {
File file = new File(args[0]);
File fileq = new File(args[1]);
File outputfile = new File(args[2]);
BufferedReader bf = new BufferedReader(new FileReader(file));
BufferedReader bfq = new BufferedReader(new FileReader(fileq));
String temp = "";
bf.readLine();
while ((temp = bf.readLine())!=null) {
seq1 += temp;
}
bfq.readLine();
while ((temp = bfq.readLine())!=null) {
seq2 += temp;
}
m = new int[seq2.length() + 1][seq1.length() + 1];
trace = new int[seq2.length() + 1][seq1.length() + 1];
m[0][0] = 0;
for (int i = 1; i <= seq2.length(); i++) {
m[i][0] = 0;
}
for (int i = 1; i <= seq1.length(); i++) {
m[0][i] = 0;
}
for (int i = 1; i <= seq2.length(); i++) {
for (int j = 1; j <= seq1.length(); j++) {
if (seq2.charAt(i - 1) == seq1.charAt(j - 1)) {
digonal = m[i - 1][j - 1] + 5;
} else {
digonal = m[i - 1][j - 1] - 4;
}
up = m[i - 1][j] - 10;
left = m[i][j - 1] - 10;
m[i][j] = Math.max(Math.max(digonal, up), left);
if (m[i][j] == digonal) {
trace[i][j] = 0;
} else if (m[i][j] == up) {
trace[i][j] = 1;
} else if (m[i][j] == left) {
trace[i][j] = 2;
}
if (m[i][j] <= 0) {
m[i][j] = 0;
}
}
}
int max = 0;
int maxi = 0, maxj = 0;
for (int i = 0; i <= seq2.length(); i++) {
for (int j = 0; j <= seq1.length(); j++) {
if (max <= m[i][j]) {
max = m[i][j];
maxi = i;
maxj = j;
}
}
}
System.out.println("max = " + max + " i = " + maxi + " j = " + maxj);
printTrace(maxi, maxj);
BufferedWriter bw = new BufferedWriter(new FileWriter(outputfile));
bw.write(seq1align);
bw.newLine();
bw.newLine();
bw.write(seq2align);
bw.flush();
bf.close();
bfq.close();
bw.close();
System.out.println("종료");
}
public static void printTrace(int i, int j) {
if (m[i][j]==0||(i == 0 && j == 0)) {
return;
}
if (trace[i][j] == 0) {
seq1align = seq1.charAt(j - 1)+seq1align;
seq2align = seq2.charAt(i - 1)+seq2align;
printTrace(i - 1, j - 1);
}
else if (trace[i][j] == 1) {
seq1align = "-"+seq1align;
seq2align = seq2.charAt(i-1)+seq2align;
printTrace(i - 1, j);
}
else if(trace[i][j] == 2){
seq1align = seq1.charAt(j-1)+seq1align;
seq2align = "-"+seq2align;
printTrace(i, j - 1);
}
else {
return;
}
}
'Algorithm' 카테고리의 다른 글
| [LeetCode] Number of Changing Keys (0) | 2024.02.24 |
|---|---|
| [LeetCode] 3042. Count Prefix and Suffix Pairs (0) | 2024.02.23 |


댓글